Section: New Results
Development of a Normal Modes Analysis SAMSON Element
Participants : Yassine Naimi, Alexandre Hoffmann, Sergei Grudinin.
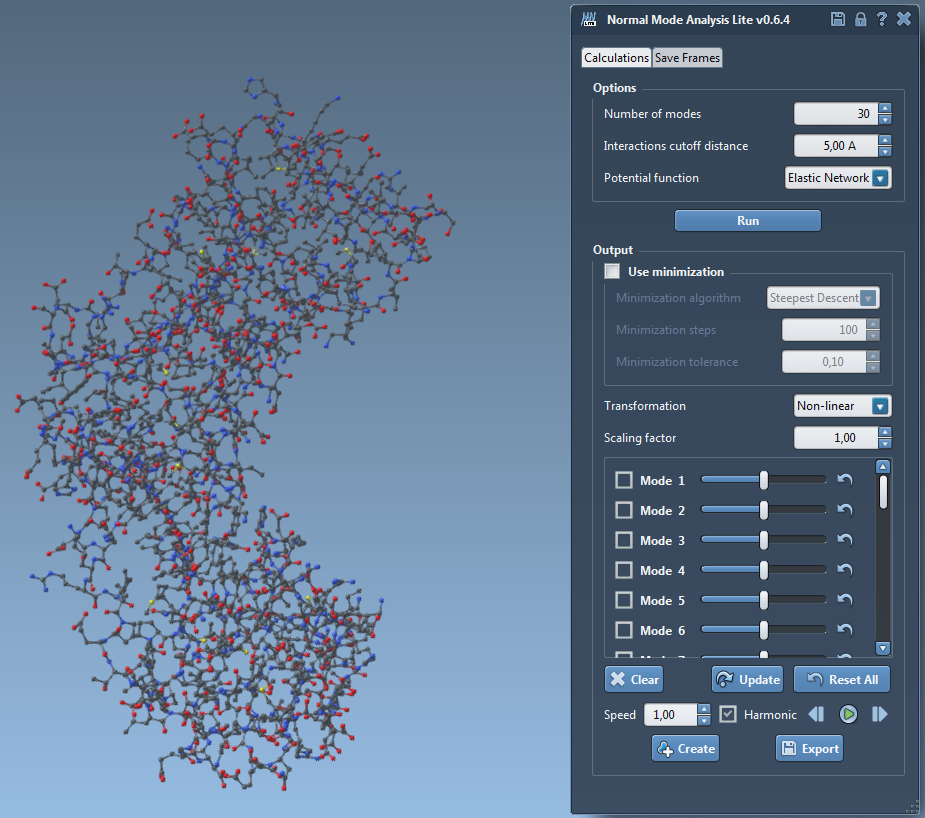
We developed a SAMSON Element based on the method proposed by Alexandre Hoffman and Sergei Grudinin on Linux and Mac operating systems. This SAMSON Element was implemented in two versions, a Lite version and an Advanced version.
The first one (14) computes the nonlinear normal modes of a molecular system (protein, RNA, DNA) very quickly using the NOLB algorithm developed by Alexandre Hoffmann and Sergei Grudinin (J. Chem. Theory Comput., 2017, 13 (5), pp 2123-2134, DOI: 10.1021 / acs.jctc.7b00197.). The user indicates the desired number of modes, the interactions cutoff distance and the potential function. For now, the elastic network model potential is the one that is available but more potential functions, like the Gaussian network model, will be added in the future. In the output, each mode is represented by a slider. The user can visualize the motion of each mode independently by moving its corresponding slider manually or by checking its checkbox and then pressing on the play button. Also, the user can visualize the motion of a combination of modes selecting them before playing the motion.
The transformations used in this motion can be set to linear or nonlinear and the amplitude of the motion can be increased/decreased by changing the scaling factor. During this motion, the user can activate a real time minimization using one of the provided algorithms (steepest descent, conjugated gradient or LBGF) and defined values of minimization steps and minimization tolerance. Finally, the user can either save/export a given conformation of the structure or the entire displayed trajectory by going into the "Save Frames" tabulation of the SAMSON element.
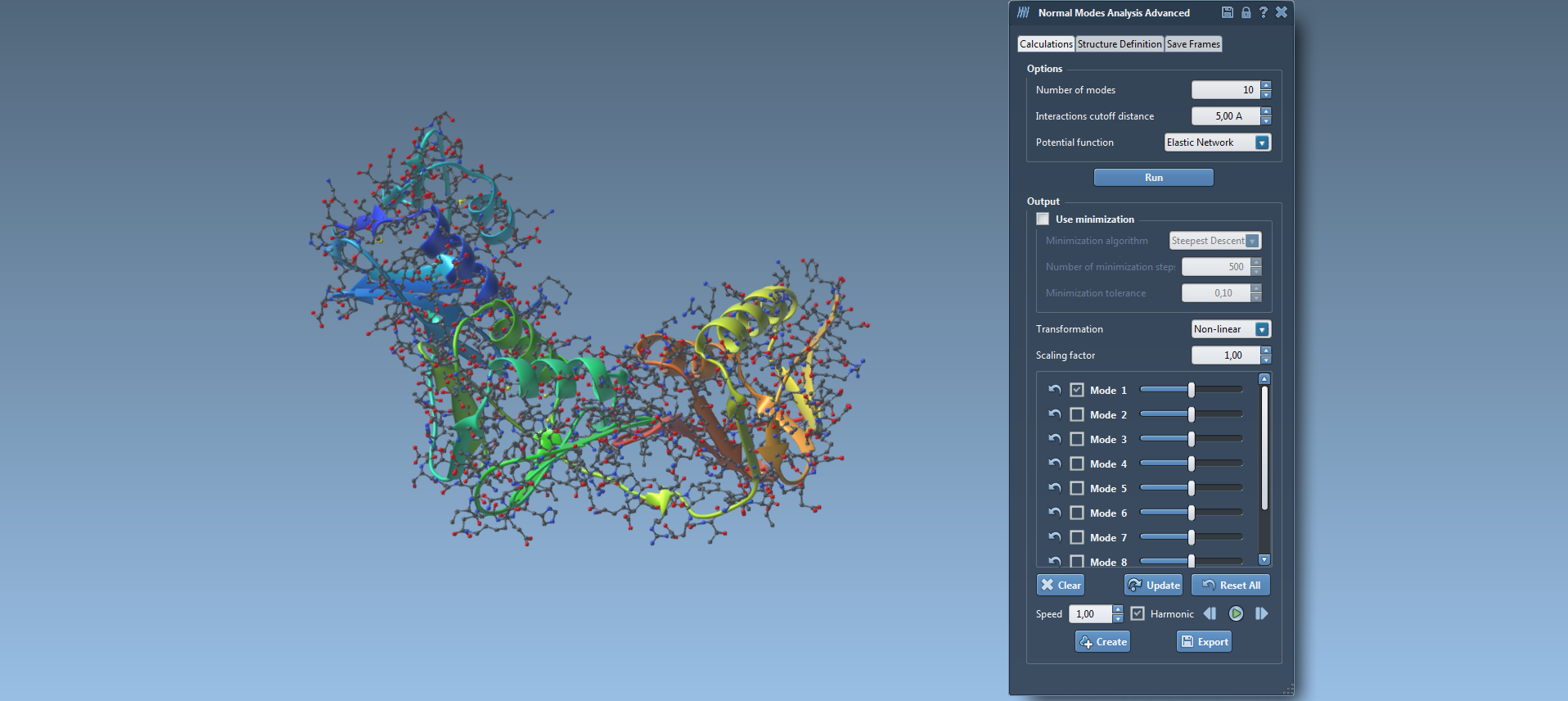
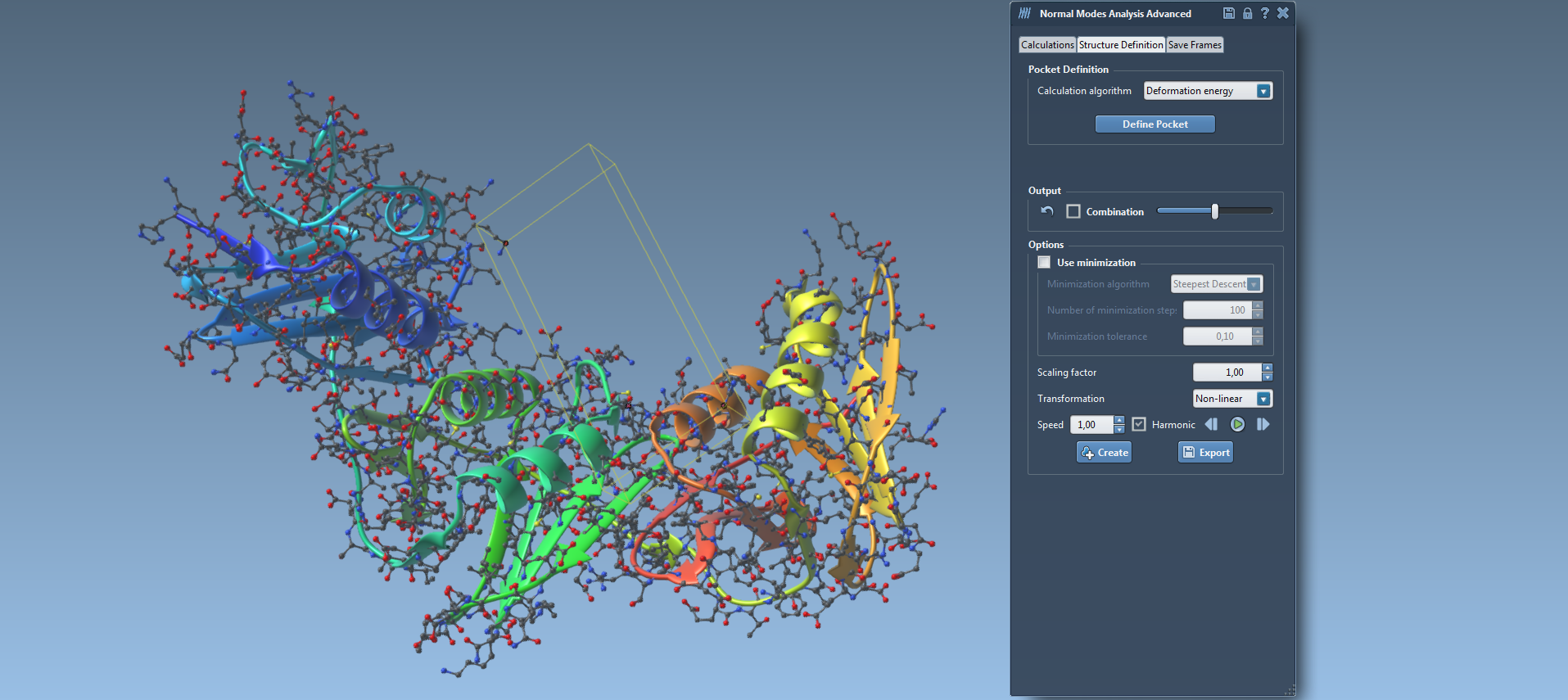
The second one (15) is an advanced version of the Nonlinear Normal Modes Analysis module. It computes the nonlinear normal modes of a molecular system (protein, RNA, DNA) in the same time given that it uses the same algorithm developed by Alexandre Hoffmann and Sergei Grudinin (J. Chem. Theory Comput., 2017, 13 (5), pp 2123-2134, DOI: 10.1021 / acs.jctc.7b00197.). It has the same functionalities than the Lite version of the Normal Modes Analysis element. In addition, the element has an additional tabulation called “Structure Definition” (16). In this section, users can define a pocket and ask for a combination of modes that contribute the most to the opening and closing of this pocket. Also, in the near future, another functionality will be added in the “Structure Definition” tabulation called “reference structure definition”. Using this functionality, users can define the conformation of structure as a reference and the element will provide a combination of modes that will lead this structure to present this conformation.